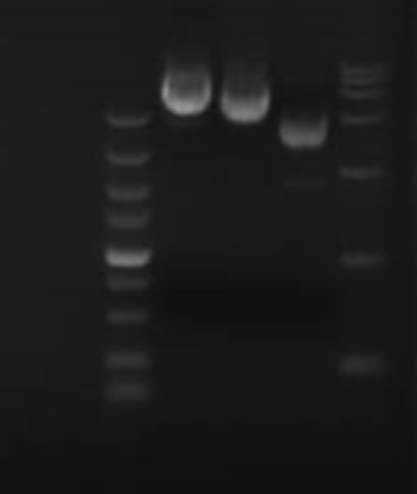
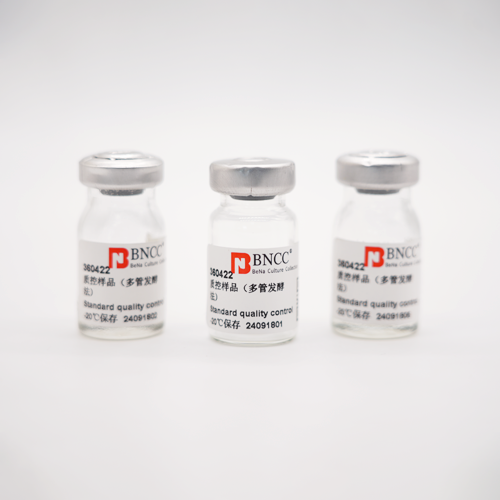
Genomic DNA
Product content: 5μg ;
Purity: (A260/A280):1.7-2.0
Product format: Freeze-dried powder
Storage conditions: 2~8 ℃
Validity period: 6 months
Biosafety level: 1
Purpose: Molecular research
Notes: If you find any abnormal conditions such as damage on the day of receipt, please contact customer service within 24 hours; after opening, it should be used up once and cannot be stored. If it is not opened temporarily, it can be stored at -20°C. Please operate in strict accordance with this instruction, otherwise it will cause abnormality, deactivation, etc., and no reissue service will be provided.
Steps:
1. Prepare sterile ddH2O;
2. centrifuge the sample tube 12000r /min for 1min, and add the corresponding volume of sterile ddH2O as required;
3. It can be used normally after a short period of centrifugation after oscillation;

 info@bncc.com
info@bncc.com
 - English
- English
 - Japanese
- Japanese